
This is a summary of Unsupervised learning by competing hidden units.
This paper introduces a novel unsupervised learning technique. There’s (almost) no backprop and the model isn’t trained for a particular task. The two authors, coming from neuroscience and computer science backgrounds based this work on two biological observations:
1- Synapses changes are local:
In biology, the synapse update depends on the activities of the presynaptic cell and the postsynaptic cell and perhaps on some global variables such as how well the task was carried out. (page 1)
The weight of a cell between A and B trained with backpropagation not only depends on the activity of A and B but also on the previous layer’s activity and the training labels. So it doesn’t depend on A, B activity but other potentially any other neurons in the network. This is inspired by Hebb‘s idea.
2- Animals learn without labeled data and fewer data than neural networks trained with backpropagation:
Second, higher animals require extensive sensory experience to tune the early […] visual system into an adult system. This experience is believed to be predominantly observational, with few or no labels, so that there is no explicit task. (page 1)
Unsupervised local training
Authors managed to train their model on MNIST and CIFAR-10 with only forward passes, meaning:
- This technique is less computationally demanding, its computational complexity is comparable to the computational complexity of the forward pass in backpropagation (source).
- Doesn’t require to train the model on a given task to make meaningful representation from the data.
The blue rectangles are the authors “biological learning algorithm”. First, the data is going through it, without any label or any indication on the task it’ll be used for. Once trained a fully connected network is appended on top of it in order to specialize the model and make the desired predictions. This part is trained using backpropagation.
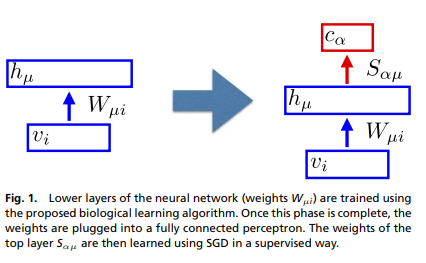
Usually to compute the activity of the hidden layer hμ, we project the input vi on it by multiplying it with a matrix Wμi and then apply non-linearity. In this new technique the hμ activity is computed solving this differential equation:
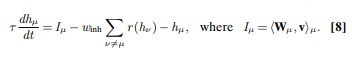
μis the index of the hidden layer we want to updateτis a timescale of the processIμis the input current- The second term, the sum of all other hidden layers, introduce competition between neurons. Stronger units will inhibit weaker ones. Without it, all neurons will fire activation when input is shown. Note that this term introduces lateral connections between units since they units within the same layer can be connected to each other.
ris a ReLU andwinhis a hyperparameter constant.
Since training is local and requires only forward passes, this architecture is different from an auto-encoder.
In action
In an experiment on MNIST and CIFAR-10, the authors trained 2000 hidden units using their biological technique to find the matrix Wμi:
- Hidden units were initialized with a normal distribution
- Hidden units are trained (again, without explicit task or labels)
- Those units are then frozen and plugged to a perceptron
- The perceptron weights were trained using SGD
The training error on MNIST can be seen in the rightmost figure of the image below (BP stands for backpropagation and BIO for the proposed approach). We can see that despite a higher training error, the testing error is very close to the model trained end-to-end.
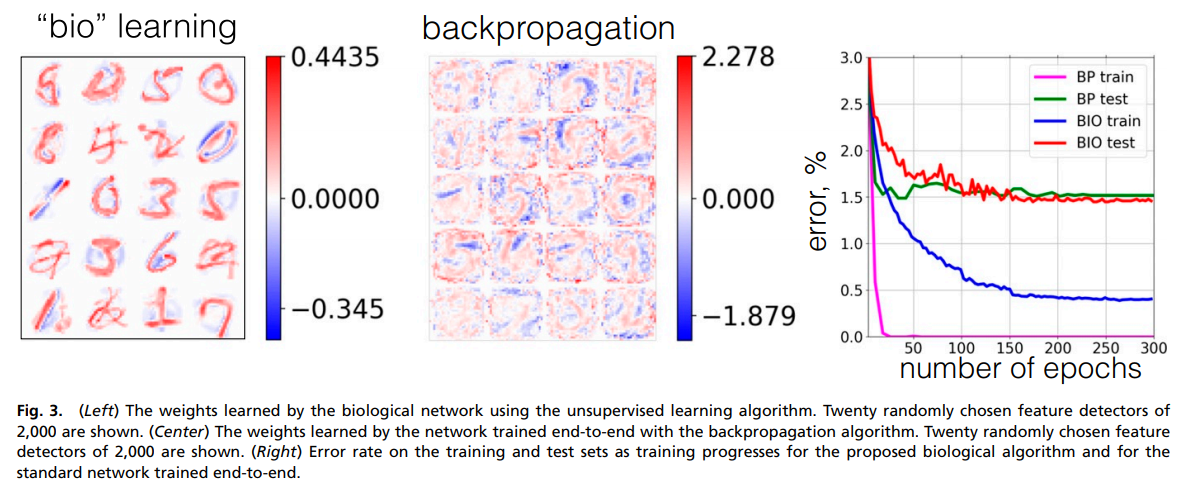
On MNIST, we can see that the features learned by the proposed biological learning algorithm (left figure) are different from the one trained with backpropagation (middle figure).
the network learns a distributed representation of the data over multiple hidden units. This representation, however, is very different from the representation learned by the network trained end-to-end, as is clear from comparison of Fig. 3, Left and Center.
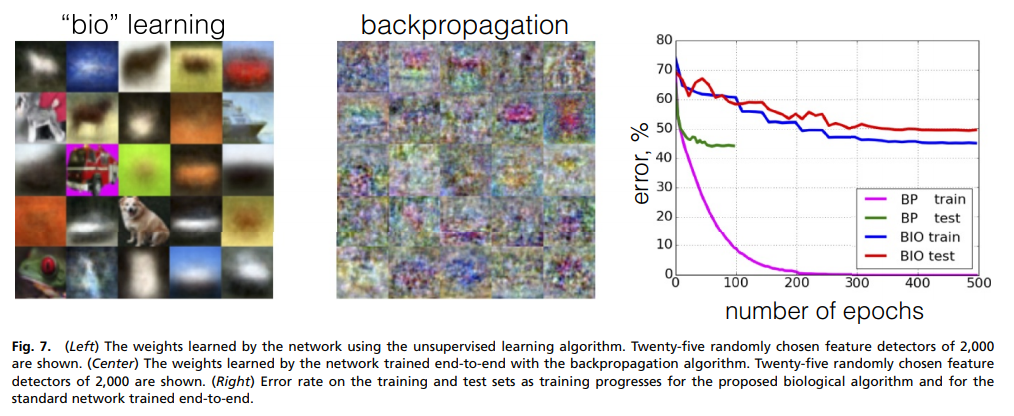
Similarly for CIFAR-10:
tldr
no top–down propagation of information, the synaptic weights are learned using only bottom-up signals, and the algorithm is agnostic about the task that the network will have to solve eventually in the top layer (page 8)
- A new unsupervised training technique, where the task isn’t defined, the training set goes through the model and is trained without backpropagation. A fully connected perceptron is appended on top, trained with backpropagation with the lower unsupervised submodel is frozen.
- This technique shows poorer but near state of the art generalizations performances on MNIST and CIFAR.
- There’s no forward/backward, each cell is potentially connected to every other, including on its own layer.
| Author | Organization | Previous work |
|---|---|---|
| Dmitry Krotov | MIT, IBM (Watson, Research), Princeton | Dense Associative Memory Is Robust to Adversarial Inputs |
| John J. Hopfield | Princeton Neuroscience Institute | same |
Complementary resources:
- Video presentation by one of the authors at MIT.
- Github for reproduction.
- Blog post on IBM’s blog.